基础知识之血氧传感器
1.什么是血氧传感器?
血氧传感器是一种用于测量人体血液中氧气饱和度的设备。它通过非侵入性或微创性的方式获取血氧水平的相关数据。血氧传感器通常使用光学原理来工作。
本文引用地址: //m.amcfsurvey.com/article/202403/456426.htm血氧传感器中最常见的类型是脉搏血氧饱和度(SpO2)传感器,也被称为脉搏血氧传感器。SpO2传感器利用光的吸收特性来测量血红蛋白的氧合程度。它通过发射两种不同波长的光(通常是红光和红外光)经过皮肤照射到血液中,然后通过相应的光电传感器测量经过皮肤反射回来的光的强度。根据红光和红外光的吸收差异,可以计算出血液中氧气的饱和度。
人体需要并调节血液中氧气的非常精确和特定的平衡。人体的正常动脉血氧饱和度(SpO2)为97-100%,或96-99%。如果该水平低于90%,则被认为是低氧血症。动脉血氧水平低于80%可能会损害器官功能,例如大脑和心脏,应及时解决。持续的低氧水平可能导致呼吸或心脏骤停。
血氧传感器常见于医疗领域,特别是在监护设备、手持式脉搏氧饱和度仪和睡眠呼吸监测等应用中。此外,它们也逐渐应用于个人健康监测设备,如智能手环、智能手表等。

图1:max30102光电式心率血氧传感器
2. 血氧传感器是如何工作的?
血氧传感器通常使用光学原理来测量血红蛋白的氧合程度,其中最常见的类型是脉搏氧血饱和度(SpO2)传感器。
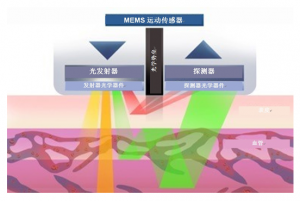
图2:光学原理
以下是血氧传感器的工作原理:
血氧传感器只能提供间接的血氧饱和度测量结果,并且有一定的误差范围。其他因素如温度、灯光干扰、运动等也可能对测量结果产生影响。
3. 如何应用血氧传感器?
1 临床监护:血氧传感器常用于临床监护中,例如在手术室、急诊室和重症监护病房。通过监测患者的血氧饱和度(SPO2)水平,医护人员可以实时了解患者的氧气供应情况,及时发现并处理低氧血症或窒息等问题,确保患者的安全。

2 睡眠呼吸监测:血氧传感器被广泛应用于睡眠呼吸监测领域。睡眠时,佩戴血氧传感器的设备(如脉搏氧饱和度仪)能够监测睡眠者的血氧水平。通过分析血氧饱和度数据,医生或睡眠专家可以评估睡眠质量、检测睡眠呼吸暂停等呼吸障碍,并为患者提供相应的治疗建议。
3 慢性阻塞性肺疾病(COPD)管理:COPD患者常使用血氧传感器来管理他们的疾病。他们可以在家中使用脉搏氧饱和度仪来测量自己的血氧水平,并追踪数据的变化。这有助于监测疾病的进展、评估治疗效果,并及时采取相应的措施,如调整药物剂量或进行氧疗。
4 家庭健康监测:现代的脉搏氧饱和度仪(Pulse Oximeter)通常集成了血氧传感器,被广泛应用于家庭健康监测场景。人们可以通过在家中使用这种设备来监测自己或家人的血氧水平,以及心率等信息。这对于早期发现可能存在的呼吸系统问题、心血管疾病和睡眠呼吸暂停等状况非常有帮助。此外,一些健康追踪设备和智能手表也集成了血氧传感器,能够提供用户的血氧饱和度数据,帮助用户更好地了解自己的健康状况。
5 高海拔登山:登山者在攀登高海拔地区时通常会面临低氧环境。血氧传感器被用于监测登山者的血氧水平,帮助他们了解身体在高海拔环境下的氧气供应情况。这样的信息可以帮助他们判断是否需要停止攀登或采取其他适当的行动来避免高山病等潜在风险。
6 运动训练和健身监测:血氧传感器在运动训练和健身监测中起着重要作用。运动员可以使用集成了血氧传感器的可穿戴设备,如智能手表或运动耳机,来监测他们的血氧水平和心率等数据。这些数据可以帮助运动员和教练员了解身体在运动过程中的氧气摄取能力和运动耐力水平,并进行相应的训练调整。此外,血氧传感器还可以帮助跑步爱好者监测自己的运动表现,提供健身指导和优化跑步计划。
4. 主要的血氧传感器供应商
Maxim Integrated :Maxim Integrated是一家知名的集成电路设计和生产公司,提供多种心率传感器芯片和模块。其中,MAX30102是一款常见的心率传感器模块,集成了红外LED、红光LED和光电传感器,适用于便携设备和健康监测设备等应用。
Texas Instruments(TI):TI是一家全球领先的半导体公司,提供多款生物传感器芯片和模块。AFE4404是TI的一款心率监测芯片,集成了红外LED、绿光LED、光电传感器和ADC等功能,可实现高精度的心率和血氧浓度测量。
Analog Devices(ADI):ADI是一家知名的模拟与数字混合信号处理技术供应商,提供多种生物传感器芯片和模块。AD8232是ADI的一款心率传感器芯片,专为心电图(ECG)采集设计,具备高性能和低功耗特点。
Nellcor (Medtronic):Nellcor是Medtronic旗下的品牌,专注于提供高质量的血氧传感器芯片和模块。他们的SpO2传感器采用专有的信号处理算法,具有高灵敏度和抗干扰能力。Nellcor的血氧传感器在医疗领域广泛应用,包括重症监护、手术室和急诊等环境。
NXP Semiconductors:NXP Semiconductors是一家全球领先的半导体解决方案提供商,他们提供适用于医疗应用的心率、血氧和心电图传感器芯片。
Silicon Labs:Silicon Labs是一家专注于集成电路解决方案的公司,他们提供用于生物传感器应用的芯片产品,包括心率、血氧和心电图传感器芯片。
5. 参考案例
在Thonny使用Mircopython编写程序控制RP2040控制Max30102读取心率数据
Max30102芯片介绍
MAX30102是一个集成的脉搏血氧计和心率监测器模块。它包括内部LED、光电探测器、光学元件和具有环境光抑制功能的低噪声电子器件。
MAX30102使用一个1.8V电源和一个用于内部LED的独立3.3V电源。通过标准I2C兼容接口进行通信。该模块可以通过零待机电流的软件关闭,使电源导轨始终保持通电状态。
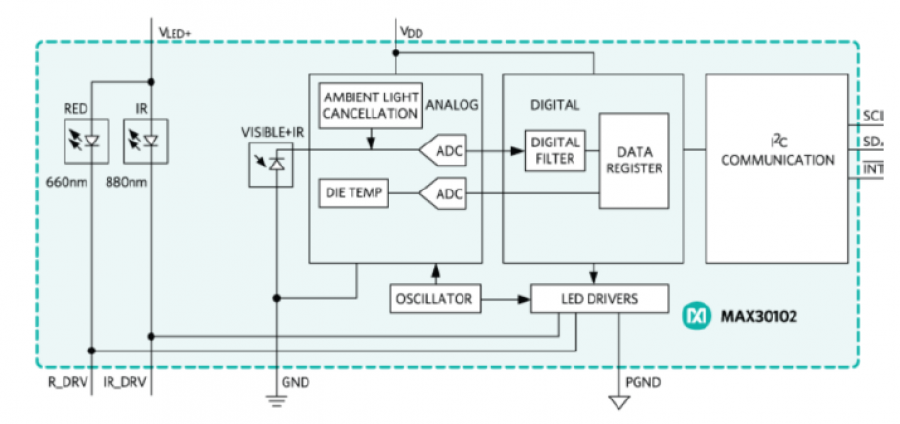
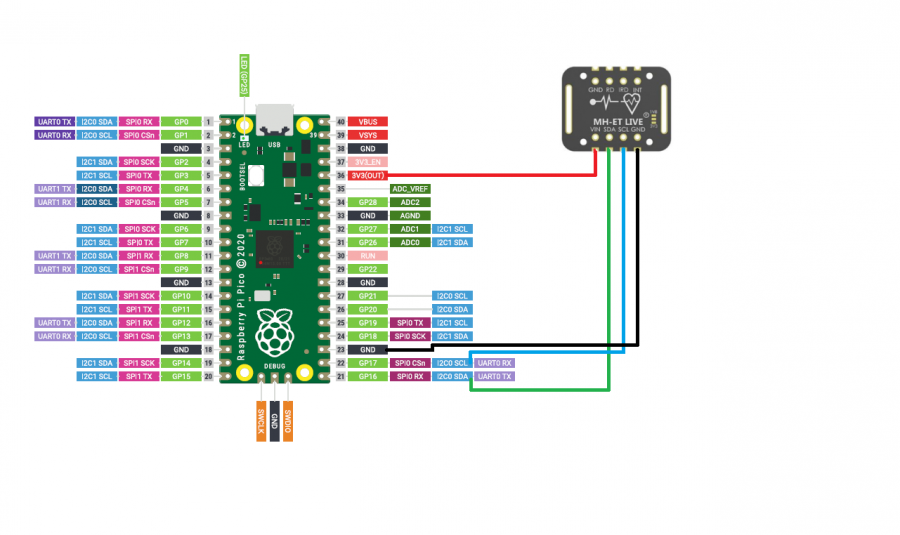
电路连接
程序代码
程序文件需要文件
前面两个文件可在github上面下载https://github.com/n-elia/MAX30102-MicroPython-driver/tree/main/max30102
spo2cal.py程序如下
# -*-coding:utf-8 # 25 samples per second (in algorithm.h)
SAMPLE_FREQ = 25
# taking moving average of 4 samples when calculating HR # in algorithm.h, "DONOT CHANGE" comment is attached
MA_SIZE = 4
# sampling frequency * 4 (in algorithm.h)
BUFFER_SIZE = 100
# this assumes ir_data and red_data as np.array
def calc_hr_and_spo2(ir_data, red_data):
"""
By detecting peaks of PPG cycle and corresponding AC/DC
of red/infra-red signal, the an_ratio for the SPO2 is computed.
"""
# get dc mean
ir_mean = int(sum(ir_data) / len(ir_data)) # remove DC mean and inver signal # this lets peak detecter detect valley
x = [ir_mean - x for x in ir_data] # 4 point moving average # x is np.array with int values, so automatically casted to int
for i in range(len(x) - MA_SIZE):
x[i] = sum(x[i:i + MA_SIZE]) / MA_SIZE
# calculate threshold
n_th = int(sum(x) / len(x))
n_th = 30 if n_th < 30 else n_th # min allowed
n_th = 60 if n_th > 60 else n_th # max allowed
ir_valley_locs, n_peaks = find_peaks(x, BUFFER_SIZE, n_th, 4, 15)
# print(ir_valley_locs[:n_peaks], ",", end="")
peak_interval_sum = 0
if n_peaks >= 2:
for i in range(1, n_peaks):
peak_interval_sum += (ir_valley_locs[i] - ir_valley_locs[i - 1])
peak_interval_sum = int(peak_interval_sum / (n_peaks - 1))
hr = int(SAMPLE_FREQ * 60 / peak_interval_sum)
hr_valid = True else:
hr = -999 # unable to calculate because # of peaks are too small
hr_valid = False
# ---------spo2--------- # find precise min near ir_valley_locs (???)
exact_ir_valley_locs_count = n_peaks
# find ir-red DC and ir-red AC for SPO2 calibration ratio # find AC/DC maximum of raw
# FIXME: needed??
for i in range(exact_ir_valley_locs_count):
if ir_valley_locs[i] > BUFFER_SIZE:
spo2 = -999 # do not use SPO2 since valley loc is out of range
spo2_valid = False return hr, hr_valid, spo2, spo2_valid
i_ratio_count = 0
ratio = [] # find max between two valley locations # and use ratio between AC component of Ir and Red DC component of Ir and Red for SpO2
red_dc_max_index = -1
ir_dc_max_index = -1
for k in range(exact_ir_valley_locs_count - 1):
red_dc_max = -16777216
ir_dc_max = -16777216
if ir_valley_locs[k + 1] - ir_valley_locs[k] > 3:
for i in range(ir_valley_locs[k], ir_valley_locs[k + 1]):
if ir_data[i] > ir_dc_max:
ir_dc_max = ir_data[i]
ir_dc_max_index = i if red_data[i] > red_dc_max:
red_dc_max = red_data[i]
red_dc_max_index = i
red_ac = int((red_data[ir_valley_locs[k + 1]] - red_data[ir_valley_locs[k]]) * (red_dc_max_index - ir_valley_locs[k]))
red_ac = red_data[ir_valley_locs[k]] + int(red_ac / (ir_valley_locs[k + 1] - ir_valley_locs[k]))
red_ac = red_data[red_dc_max_index] - red_ac # subtract linear DC components from raw
ir_ac = int((ir_data[ir_valley_locs[k + 1]] - ir_data[ir_valley_locs[k]]) * (ir_dc_max_index - ir_valley_locs[k]))
ir_ac = ir_data[ir_valley_locs[k]] + int(ir_ac / (ir_valley_locs[k + 1] - ir_valley_locs[k]))
ir_ac = ir_data[ir_dc_max_index] - ir_ac # subtract linear DC components from raw
nume = red_ac * ir_dc_max
denom = ir_ac * red_dc_max if (denom > 0 and i_ratio_count < 5) and nume != 0:
# original cpp implementation uses overflow intentionally. # but at 64-bit OS, Pyhthon 3.X uses 64-bit int and nume*100/denom does not trigger overflow # so using bit operation ( &0xffffffff ) is needed
ratio.append(int(((nume * 100) & 0xffffffff) / denom))
i_ratio_count += 1 # choose median value since PPG signal may vary from beat to beat
ratio = sorted(ratio) # sort to ascending order
mid_index = int(i_ratio_count / 2)
ratio_ave = 0
if mid_index > 1:
ratio_ave = int((ratio[mid_index - 1] + ratio[mid_index]) / 2)
else:
if len(ratio) != 0:
ratio_ave = ratio[mid_index] # why 184?
# print("ratio average: ", ratio_ave)
if ratio_ave > 2 and ratio_ave < 184:
# -45.060 * ratioAverage * ratioAverage / 10000 + 30.354 * ratioAverage / 100 + 94.845
spo2 = -45.060 * (ratio_ave ** 2) / 10000.0 + 30.054 * ratio_ave / 100.0 + 94.845
spo2_valid = True else:
spo2 = -999
spo2_valid = False
return hr - 20, hr_valid, spo2, spo2_valid
def find_peaks(x, size, min_height, min_dist, max_num):
"""
Find at most MAX_NUM peaks above MIN_HEIGHT separated by at least MIN_DISTANCE
"""
ir_valley_locs, n_peaks = find_peaks_above_min_height(x, size, min_height, max_num)
ir_valley_locs, n_peaks = remove_close_peaks(n_peaks, ir_valley_locs, x, min_dist)
n_peaks = min([n_peaks, max_num]) return ir_valley_locs, n_peaks
def find_peaks_above_min_height(x, size, min_height, max_num):
"""
Find all peaks above MIN_HEIGHT
"""
i = 0
n_peaks = 0
ir_valley_locs = [] # [0 for i in range(max_num)]
while i < size - 1:
if x[i] > min_height and x[i] > x[i - 1]: # find the left edge of potential peaks
n_width = 1
# original condition i+n_width < size may cause IndexError # so I changed the condition to i+n_width < size - 1
while i + n_width < size - 1 and x[i] == x[i + n_width]: # find flat peaks
n_width += 1
if x[i] > x[i + n_width] and n_peaks < max_num: # find the right edge of peaks # ir_valley_locs[n_peaks] = i
ir_valley_locs.append(i)
n_peaks += 1 # original uses post increment
i += n_width + 1
else:
i += n_width else:
i += 1 return ir_valley_locs, n_peaks
def remove_close_peaks(n_peaks, ir_valley_locs, x, min_dist):
"""
Remove peaks separated by less than MIN_DISTANCE
""" # should be equal to maxim_sort_indices_descend # order peaks from large to small
# should ignore index:0
sorted_indices = sorted(ir_valley_locs, key=lambda i: x[i])
sorted_indices.reverse() # this "for" loop expression does not check finish condition # for i in range(-1, n_peaks):
i = -1
while i < n_peaks:
old_n_peaks = n_peaks
n_peaks = i + 1
# this "for" loop expression does not check finish condition # for j in (i + 1, old_n_peaks):
j = i + 1
while j < old_n_peaks:
n_dist = (sorted_indices[j] - sorted_indices[i]) if i != -1 else (sorted_indices[j] + 1) # lag-zero peak of autocorr is at index -1
if n_dist > min_dist or n_dist < -1 * min_dist:
sorted_indices[n_peaks] = sorted_indices[j]
n_peaks += 1 # original uses post increment
j += 1
i += 1
sorted_indices[:n_peaks] = sorted(sorted_indices[:n_peaks]) return sorted_indices, n_peaks
if __name__ == "__main__":
hr, hrb, sp, spb = calc_hr_and_spo2([12853, 15573, 15580, 15586, 15587, 15567, 15520, 15480, 15464, 15460, 15462, 15466, 15473, 15479, 15485, 15490, 15495, 15503, 15512, 15518, 15521, 15521, 15518, 15517, 15522, 15527, 15536, 15547, 15558, 15568, 15577, 15587, 15594, 15604, 15610, 15616, 15620, 15624, 15625, 15615, 15576, 15531, 15508, 15500, 15502, 15509, 15516, 15523, 15528, 15533, 15538, 15547, 15556, 15564, 15564, 15560, 15556, 15556, 15559, 15564, 15570, 15579, 15588, 15599, 15610, 15619, 15628, 15635, 15642, 15649, 15655, 15662, 15669, 15672, 15661, 15621, 15571, 15546, 15537, 15538, 15545, 15553, 15560, 15565, 15570, 15577, 15585, 15593, 15600, 15601, 15597, 15592, 15591, 15594, 15600, 15608, 15617, 15626, 15633, 15640], [12258, 14318, 14322, 14324, 14326, 14317, 14299, 14284, 14280, 14279, 14280, 14283, 14285, 14288, 14292, 14294, 14297, 14299, 14302, 14304, 14305, 14305, 14304, 14304, 14306, 14308, 14311, 14316, 14321, 14325, 14329, 14333, 14329, 14329, 14332, 14335, 14336, 14338, 14338, 14333, 14315, 14295, 14286, 14283, 14285, 14288, 14292, 14295, 14297, 14298, 14301, 14305, 14309, 14312, 14312, 14310, 14308, 14308, 14309, 14312, 14315, 14318, 14322, 14327, 14332, 14336, 14341, 14344, 14347, 14350, 14351, 14354, 14357, 14359, 14353, 14335, 14313, 14304, 14300, 14302, 14305, 14309, 14312, 14314, 14316, 14319, 14323, 14326, 14329, 14329, 14326, 14325, 14324, 14326, 14328, 14332, 14336, 14341, 14345, 14349])
print("hr detected:", hrb)
print("sp detected:", spb) if (hrb == True and hr != -999):
hr2 = int(hr)
print("Heart Rate : ", hr2)
if (spb == True and sp != -999):
sp2 = int(sp)
print("SPO2 : ", sp2)
HR_SpO2.py程序如下:
from machine import SoftI2C, Pin, Timer
from utime import ticks_diff, ticks_us
from max30102 import MAX30102, MAX30105_PULSE_AMP_MEDIUM
from spo2cal import calc_hr_and_spo2
BEATS = 0 # 存储心率
FINGER_FLAG = False # 默认表示未检测到手指
SPO2 = 0 # 存储血氧
TEMPERATURE = 0 # 存储温度
def display_info(t):
# 如果没有检测到手指,那么就不显示 if FINGER_FLAG is False:
return
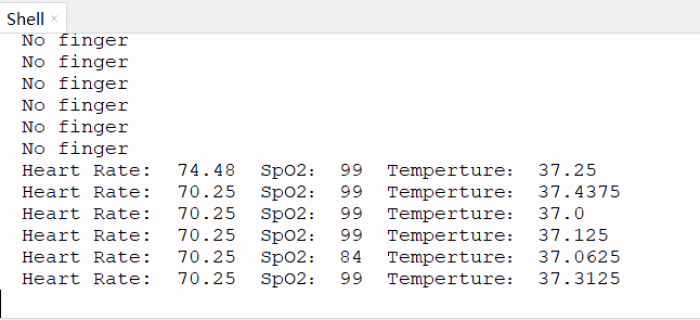
print('Heart Rate: ', BEATS, " SpO2:", SPO2, " Temperture:", TEMPERATURE)
def main():
global BEATS, FINGER_FLAG, SPO2, TEMPERATURE # 如果需要对全局变量修改,则需要global声明
# 创建I2C对象(检测MAX30102)
i2c = SoftI2C(sda=Pin(16), scl=Pin(17), freq=400000) # Fast: 400kHz, slow: 100kHz
# 创建传感器对象
sensor = MAX30102(i2c=i2c) # 检测是否有传感器 if sensor.i2c_address not in i2c.scan():
print("没有找到传感器")
return
elif not (sensor.check_part_id()):
# 检查传感器是否兼容
print("检测到的I2C设备不是MAX30102或者MAX30105")
return
else:
print("传感器已识别到") # 配置
sensor.setup_sensor()
sensor.set_sample_rate(400)
sensor.set_fifo_average(8)
sensor.set_active_leds_amplitude(MAX30105_PULSE_AMP_MEDIUM)
t_start = ticks_us() # Starting time of the acquisition
MAX_HISTORY = 32
history = []
beats_history = []
beat = False
red_list = []
ir_list = [] while True:
sensor.check()
if sensor.available():
# FIFO 先进先出,从队列中取数据。都是整形int
red_reading = sensor.pop_red_from_storage()
ir_reading = sensor.pop_ir_from_storage() if red_reading < 1000:
print('No finger')
FINGER_FLAG = False # 表示没有放手指 continue
else:
FINGER_FLAG = True # 表示手指已放
# 计算心率
history.append(red_reading) # 为了防止列表过大,这里取列表的后32个元素
history = history[-MAX_HISTORY:] # 提取必要数据
minima, maxima = min(history), max(history)
threshold_on = (minima + maxima * 3) // 4 # 3/4
threshold_off = (minima + maxima) // 2 # 1/2 if not beat and red_reading > threshold_on:
beat = True
t_us = ticks_diff(ticks_us(), t_start)
t_s = t_us/1000000
f = 1/t_s
bpm = f * 60
if bpm < 500:
t_start = ticks_us()
beats_history.append(bpm)
beats_history = beats_history[-MAX_HISTORY:] # 只保留最大30个元素数据
BEATS = round(sum(beats_history)/len(beats_history), 2) # 四舍五入 if beat and red_reading < threshold_off:
beat = False
# 计算血氧
red_list.append(red_reading)
ir_list.append(ir_reading)
# 最多 只保留最新的100个
red_list = red_list[-100:]
ir_list = ir_list[-100:]
# 计算血氧值 if len(red_list) == 100 and len(ir_list) == 100:
hr, hrb, sp, spb = calc_hr_and_spo2(red_list, ir_list)
if hrb is True and spb is True:
if sp != -999:
SPO2 = int(sp) # 计算温度
TEMPERATURE = sensor.read_temperature()
if __name__ == '__main__':
tim = Timer(period=1000, mode=Timer.PERIODIC, callback=display_info)
main()
运行效果




评论